Chapter 4 Data Manipulation
4.1 Transforming Data
4.1.1 Exploring data
Useful verbs:
glimpse(): similar tostr().select(): select columns.select(var_name1, var_name2,...)filter(): select rows. For multiple conditions, can separate by:1. comma
filter(con1, con2,...)2. logic operator
&or|,filter(con1 & con2)3.
%in%: used to filter for multiple values.filter(var %in% c("var_value1", "var_value2"))arrange(): sort by variables.mutate(): change exist or create new variables.
library(tidyverse)## ── Attaching core tidyverse packages ─────────
## ✔ forcats 1.0.0 ✔ stringr 1.5.0
## ✔ lubridate 1.9.2 ✔ tibble 3.2.1
## ✔ purrr 1.0.1 ✔ tidyr 1.3.0
## ✔ readr 2.1.4
## ── Conflicts ──────── tidyverse_conflicts() ──
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()
## ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorscounties <- read_csv("data/counties.csv")## Rows: 3138 Columns: 40
## ── Column specification ──────────────────────
## Delimiter: ","
## chr (4): state, county, region, metro
## dbl (36): census_id, population, men, women, hispanic, white, black, native,...
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.glimpse(counties)## Rows: 3,138
## Columns: 40
## $ census_id <dbl> 1001, 1003, 1005, 1007, 1009, 1011, 1013, 1015, 101…
## $ state <chr> "Alabama", "Alabama", "Alabama", "Alabama", "Alabam…
## $ county <chr> "\"Autauga\"", "\"Baldwin\"", "\"Barbour\"", "\"Bib…
## $ region <chr> "South", "South", "South", "South", "South", "South…
## $ metro <chr> "Metro", "Metro", "Nonmetro", "Metro", "Metro", "No…
## $ population <dbl> 55221, 195121, 26932, 22604, 57710, 10678, 20354, 1…
## $ men <dbl> 26700, 95300, 14500, 12100, 28500, 5660, 9500, 5630…
## $ women <dbl> 28500, 99800, 12400, 10500, 29200, 5020, 10900, 604…
## $ hispanic <dbl> 2.6, 4.5, 4.6, 2.2, 8.6, 4.4, 1.2, 3.5, 0.4, 1.5, 7…
## $ white <dbl> 75.8, 83.1, 46.2, 74.5, 87.9, 22.2, 53.3, 73.0, 57.…
## $ black <dbl> 18.5, 9.5, 46.7, 21.4, 1.5, 70.7, 43.8, 20.3, 40.3,…
## $ native <dbl> 0.4, 0.6, 0.2, 0.4, 0.3, 1.2, 0.1, 0.2, 0.2, 0.6, 0…
## $ asian <dbl> 1.0, 0.7, 0.4, 0.1, 0.1, 0.2, 0.4, 0.9, 0.8, 0.3, 0…
## $ pacific <dbl> 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0…
## $ citizens <dbl> 40725, 147695, 20714, 17495, 42345, 8057, 15581, 88…
## $ income <dbl> 51281, 50254, 32964, 38678, 45813, 31938, 32229, 41…
## $ income_err <dbl> 2391, 1263, 2973, 3995, 3141, 5884, 1793, 925, 2949…
## $ income_per_cap <dbl> 24974, 27317, 16824, 18431, 20532, 17580, 18390, 21…
## $ income_per_cap_err <dbl> 1080, 711, 798, 1618, 708, 2055, 714, 489, 1366, 15…
## $ poverty <dbl> 12.9, 13.4, 26.7, 16.8, 16.7, 24.6, 25.4, 20.5, 21.…
## $ child_poverty <dbl> 18.6, 19.2, 45.3, 27.9, 27.2, 38.4, 39.2, 31.6, 37.…
## $ professional <dbl> 33.2, 33.1, 26.8, 21.5, 28.5, 18.8, 27.5, 27.3, 23.…
## $ service <dbl> 17.0, 17.7, 16.1, 17.9, 14.1, 15.0, 16.6, 17.7, 14.…
## $ office <dbl> 24.2, 27.1, 23.1, 17.8, 23.9, 19.7, 21.9, 24.2, 26.…
## $ construction <dbl> 8.6, 10.8, 10.8, 19.0, 13.5, 20.1, 10.3, 10.5, 11.5…
## $ production <dbl> 17.1, 11.2, 23.1, 23.7, 19.9, 26.4, 23.7, 20.4, 24.…
## $ drive <dbl> 87.5, 84.7, 83.8, 83.2, 84.9, 74.9, 84.5, 85.3, 85.…
## $ carpool <dbl> 8.8, 8.8, 10.9, 13.5, 11.2, 14.9, 12.4, 9.4, 11.9, …
## $ transit <dbl> 0.1, 0.1, 0.4, 0.5, 0.4, 0.7, 0.0, 0.2, 0.2, 0.2, 0…
## $ walk <dbl> 0.5, 1.0, 1.8, 0.6, 0.9, 5.0, 0.8, 1.2, 0.3, 0.6, 1…
## $ other_transp <dbl> 1.3, 1.4, 1.5, 1.5, 0.4, 1.7, 0.6, 1.2, 0.4, 0.7, 1…
## $ work_at_home <dbl> 1.8, 3.9, 1.6, 0.7, 2.3, 2.8, 1.7, 2.7, 2.1, 2.5, 1…
## $ mean_commute <dbl> 26.5, 26.4, 24.1, 28.8, 34.9, 27.5, 24.6, 24.1, 25.…
## $ employed <dbl> 23986, 85953, 8597, 8294, 22189, 3865, 7813, 47401,…
## $ private_work <dbl> 73.6, 81.5, 71.8, 76.8, 82.0, 79.5, 77.4, 74.1, 85.…
## $ public_work <dbl> 20.9, 12.3, 20.8, 16.1, 13.5, 15.1, 16.2, 20.8, 12.…
## $ self_employed <dbl> 5.5, 5.8, 7.3, 6.7, 4.2, 5.4, 6.2, 5.0, 2.8, 7.9, 4…
## $ family_work <dbl> 0.0, 0.4, 0.1, 0.4, 0.4, 0.0, 0.2, 0.1, 0.0, 0.5, 0…
## $ unemployment <dbl> 7.6, 7.5, 17.6, 8.3, 7.7, 18.0, 10.9, 12.3, 8.9, 7.…
## $ land_area <dbl> 594, 1590, 885, 623, 645, 623, 777, 606, 597, 554, …counties %>%
# Select the five columns
select(state, county, population, men, women) %>%
# Add the proportion_men variable
mutate(proportion_men = men / population) %>%
# Filter for population of at least 10,000
filter(population >= 10000, state == "California") %>%
# Arrange proportion of men in descending order
arrange(desc(proportion_men))## # A tibble: 55 × 6
## state county population men women proportion_men
## <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 California "\"Lassen\"" 32645 21800 10800 0.668
## 2 California "\"Kings\"" 150998 84000 67000 0.556
## 3 California "\"Del Norte\"" 27788 15400 12400 0.554
## 4 California "\"Amador\"" 36995 20000 17000 0.541
## 5 California "\"Tuolumne\"" 54079 28200 25900 0.521
## 6 California "\"Colusa\"" 21396 11100 10300 0.519
## 7 California "\"Mono\"" 14146 7310 6840 0.517
## 8 California "\"Trinity\"" 13373 6880 6500 0.514
## 9 California "\"Kern\"" 865736 445000 421000 0.514
## 10 California "\"Imperial\"" 178206 91200 87000 0.512
## # ℹ 45 more rows4.2 Aggregating Data
4.2.1 Count
One way we can aggregate data is to count(): to find out the number of observations.
Count variable and sort:
count(var, sort = TRUE)counties_selected <- counties %>%
select(county, region, state, population, citizens)
# Use count to find the number of counties in each region
counties_selected %>%
count(region, sort = TRUE)## # A tibble: 3 × 2
## region n
## <chr> <int>
## 1 South 1420
## 2 North~ 1271
## 3 West 447Add weight:
count(var1, wt = var2, sort = TRUE)counties_selected <- counties %>%
select(county, region, state, population, citizens)
# Find number of counties per state, weighted by citizens, sorted in descending order
counties_selected %>%
count(state, wt = citizens, sort = TRUE)## # A tibble: 50 × 2
## state n
## <chr> <dbl>
## 1 California 24280349
## 2 Texas 16864864
## 3 Florida 13933052
## 4 New York 13531404
## 5 Pennsylvan 9710416
## 6 Illinois 8979999
## 7 Ohio 8709050
## 8 Michigan 7380136
## 9 North Caro 7107998
## 10 Georgia 6978660
## # ℹ 40 more rowsMutating and counting:
“What are the US states where the most people walk to work?”
counties_selected <- counties %>%
select(county, region, state, population, walk)
counties_selected %>%
# Add population_walk containing the total number of people who walk to work
mutate(population_walk = walk * population / 100) %>%
# Count weighted by the new column, sort in descending order
count(state, wt = population_walk, sort = TRUE)## # A tibble: 50 × 2
## state n
## <chr> <dbl>
## 1 New York 1237938.
## 2 California 1017964.
## 3 Pennsylvan 505397.
## 4 Texas 430783.
## 5 Illinois 400346.
## 6 Massachuse 316765.
## 7 Florida 284723.
## 8 New Jersey 273047.
## 9 Ohio 266911.
## 10 Washington 239764.
## # ℹ 40 more rows4.2.2 group_by, summarize, ungroup
summarize(): takes many observations and turns them into one observation. Also can define multiple variables in asummarizecall.group_by(): aggregate within groups. We can group by multiple columns by passing multiple column names togroup_by.When you group by multiple columns and then summarize, it’s important to remember that the summarize “peels off” one of the groups, but leaves the rest on. For example, if you
group_by(X, Y)then summarize, the result will still be grouped byX.ungroup(): If you don’t want to keep variable as a group, you can add anotherungroup().
Summarize both population and land area by state, with the purpose of finding the density (in people per square miles).
counties %>%
select(state, county, population, land_area) %>%
# Group by state
group_by(state) %>%
# Find the total area and population
summarize(total_area = sum(land_area),
total_population = sum(population)) %>%
# Add a density column
mutate(density = total_population / total_area) %>%
# Sort by density in descending order
arrange(desc(density))## # A tibble: 50 × 4
## state total_area total_population density
## <chr> <dbl> <dbl> <dbl>
## 1 New Jersey 7356. 8904413 1210.
## 2 Rhode Isla 1034. 1053661 1019.
## 3 Massachuse 7800. 6705586 860.
## 4 Connecticu 4843 3593222 742.
## 5 Maryland 9707. 5930538 611.
## 6 Delaware 1948 926454 476.
## 7 New York 47127. 19673174 417.
## 8 Florida 53627 19645772 366.
## 9 Pennsylvan 44740 12779559 286.
## 10 Ohio 40855 11575977 283.
## # ℹ 40 more rowsYou can group by multiple columns instead of grouping by one.Summarizing by state and region:
counties %>%
select(region, state, county, population) %>%
# Group and summarize to find the total population
group_by(region, state) %>%
summarize(total_pop = sum(population)) %>%
# Calculate the average_pop and median_pop columns
summarize(average_pop = mean(total_pop),
median_pop = median(total_pop))## `summarise()` has grouped output by 'region'.
## You can override using the `.groups`
## argument.## # A tibble: 3 × 3
## region average_pop median_pop
## <chr> <dbl> <dbl>
## 1 North~ 5881989. 5419171
## 2 South 7370486 4804098
## 3 West 5722755. 27986364.2.3 slice_min, slice_max
slice_max() operates on a grouped table, and returns the largest observations in each group. The function takes two arguments: the column we want to base the ordering on, and the number of observations to extract from each group, specified with the n argument.
# The largest observations
slice_max(var, n = 1)
# The 3 largest observations
slice_max(var, n = 3)Similarly, slice_min() returns the smallest observations in each group.
# The smallest observations
slice_min(var, n = 1)
# The 3 smallest observations
slice_min(var, n = 3)The slicing are often used when creating visualizations, where we may want to highlight the extreme observations on the plot.
Q: In how many states do more people live in metro areas than non-metro areas?
counties %>%
select(state, metro, population) %>%
# Find the total population for each combination of state and metro
group_by(state, metro) %>%
summarize(total_pop = sum(population)) %>%
# Extract the most populated row for each state
slice_max(total_pop, n = 1) %>%
# Count the states with more people in Metro or Nonmetro areas
ungroup() %>%
count(metro, sort = TRUE)## `summarise()` has grouped output by 'state'.
## You can override using the `.groups`
## argument.## # A tibble: 2 × 2
## metro n
## <chr> <int>
## 1 Metro 44
## 2 Nonmetro 64.3 Selecting and Transforming Data
4.3.1 Select
4.3.1.1 Range
Select a range of columns.
select(var1, var3:var6)
# For example
select(state, county, drive:work_at_home)counties %>%
# Select state, county, population, and industry-related columns
select(state, county, population, professional:production) %>%
# Arrange service in descending order
arrange(desc(service))## # A tibble: 3,138 × 8
## state county population professional service office construction production
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Missis… "i … 10477 23.9 36.6 21.5 3.5 14.5
## 2 Texas "\"Ki… 3577 30 36.5 11.6 20.5 1.3
## 3 Texas "\"Ke… 565 24.9 34.1 20.5 20.5 0
## 4 New Yo… "\"Br… 1428357 24.3 33.3 24.2 7.1 11
## 5 Texas "\"Br… 7221 19.6 32.4 25.3 11.1 11.5
## 6 Colora… "\"Fr… 46809 26.6 32.2 22.8 10.7 7.6
## 7 Texas "\"Cu… 2296 20.1 32.2 24.2 15.7 7.8
## 8 Califo… "\"De… 27788 33.9 31.5 18.8 8.9 6.8
## 9 Minnes… "\"Ma… 5496 26.8 31.5 18.7 13.1 9.9
## 10 Virgin… "\"La… 11129 30.3 31.2 22.8 8.1 7.6
## # ℹ 3,128 more rows4.3.1.2 Contains
Specify criteria for choosing columns.
select(var1, var3, contains("char"))
# For example
select(state, county, contains("work"))4.3.1.3 Starts with
To select only the columns that start with a particular prefix.
select(var1, var3, starts_with("char"))
# For example
select(state, county, starts_with("income"))4.3.1.4 Ends with
Finds columns ending in a string.
select(var1, var3, ends_with("char"))
# For example
select(state, county, ends_with("tion"))counties %>%
# Select the state, county, population, and those ending with "work"
select(state, county, population, ends_with("work")) %>%
# Filter for counties that have at least 50% of people engaged in public work
filter(public_work > 50)## # A tibble: 7 × 6
## state county population private_work public_work family_work
## <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Alaska "\"Lake and Penins… 1474 42.2 51.6 0.2
## 2 Alaska "\"Yukon-Koyukuk C… 5644 33.3 61.7 0
## 3 California "\"Lassen\"" 32645 42.6 50.5 0.1
## 4 Hawaii "\"Kalawao\"" 85 25 64.1 0
## 5 North Dako "ta \"Sioux\"" 4380 32.9 56.8 0.1
## 6 South Dako "ta \"Todd\"" 9942 34.4 55 0.8
## 7 Wisconsin "\"Menominee\"" 4451 36.8 59.1 0.44.3.2 Rename
Often, rather than only selecting columns, we’ll sometimes want to rename the ones we already have.
Compare these two ways:
- Select
When select variables, change variables’ name at the same time.
select(var1, var2, var_newname = var_oldname)
# For example
counties %>%
select(state, county, population, unemployment_rate = unemployment)counties %>%
# Select state, county, and poverty as poverty_rate
select(state, county, poverty_rate = poverty)## # A tibble: 3,138 × 3
## state county poverty_rate
## <chr> <chr> <dbl>
## 1 Alabama "\"Autauga\"" 12.9
## 2 Alabama "\"Baldwin\"" 13.4
## 3 Alabama "\"Barbour\"" 26.7
## 4 Alabama "\"Bibb\"" 16.8
## 5 Alabama "\"Blount\"" 16.7
## 6 Alabama "\"Bullock\"" 24.6
## 7 Alabama "\"Butler\"" 25.4
## 8 Alabama "\"Calhoun\"" 20.5
## 9 Alabama "\"Chambers\"" 21.6
## 10 Alabama "\"Cherokee\"" 19.2
## # ℹ 3,128 more rows- Rename
rename() is often useful for changing the name of a column that comes out of another verb.
rename(var_newname = var_oldname)
# For example
counties %>%
select(state, county, population, unemployment) %>%
rename(unemployment_rate = unemployment)counties %>%
# Count the number of counties in each state
count(state)## # A tibble: 50 × 2
## state n
## <chr> <int>
## 1 Alabama 67
## 2 Alaska 28
## 3 Arizona 15
## 4 Arkansas 75
## 5 California 58
## 6 Colorado 64
## 7 Connecticu 8
## 8 Delaware 3
## 9 Florida 67
## 10 Georgia 159
## # ℹ 40 more rowscounties %>%
# Count the number of counties in each state
count(state) %>%
# Rename the n column to num_counties
rename(num_counties = n)## # A tibble: 50 × 2
## state num_counties
## <chr> <int>
## 1 Alabama 67
## 2 Alaska 28
## 3 Arizona 15
## 4 Arkansas 75
## 5 California 58
## 6 Colorado 64
## 7 Connecticu 8
## 8 Delaware 3
## 9 Florida 67
## 10 Georgia 159
## # ℹ 40 more rows4.3.3 Transmute
Transmute is like a combination of select and mutate: it returns a subset of the columns like select, but it can also transform and change the columns, like mutate, at the same time.
It control which variables you keep, which variables you calculate, and which variables you drop.
transmute(var1, var2, var_new = var_old*do caculate)
#For example
counties %>%
transmute(state, county, fraction_men = men / population)counties %>%
# Keep the state, county, and populations columns, and add a density column
transmute(state, county, population, density = population/land_area) %>%
# Filter for counties with a population greater than one million
filter(population > 1000000) %>%
# Sort density in ascending order
arrange(density)## # A tibble: 41 × 4
## state county population density
## <chr> <chr> <dbl> <dbl>
## 1 California "\"San Bernardino\"" 2094769 104.
## 2 Nevada "\"Clark\"" 2035572 258.
## 3 California "\"Riverside\"" 2298032 319.
## 4 Arizona "\"Maricopa\"" 4018143 437.
## 5 Florida "\"Palm Beach\"" 1378806 700.
## 6 California "\"San Diego\"" 3223096 766.
## 7 Washington "\"King\"" 2045756 967.
## 8 Texas "\"Travis\"" 1121645 1133.
## 9 Florida "\"Hillsborough\"" 1302884 1277.
## 10 Florida "\"Orange\"" 1229039 1361.
## # ℹ 31 more rowsSummary
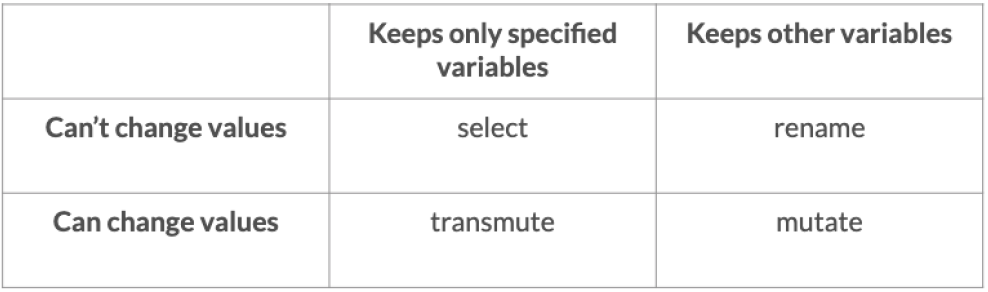
# Change the name of the unemployment column
counties %>%
rename(unemployment_rate = unemployment)
# Keep the state and county columns, and the columns containing poverty
counties %>%
select(state, county, contains("poverty"))
# Calculate the fraction_women column without dropping the other columns
counties %>%
mutate(fraction_women = women / population)
# Keep only the state, county, and employment_rate columns
counties %>%
transmute(state, county, employment_rate = employed / population)4.4 Case Study: The babynames Dataset
4.4.1 Load dataset
library(babynames)## Warning: package 'babynames' was built under R version 4.3.1babynames## # A tibble: 1,924,665 × 5
## year sex name n prop
## <dbl> <chr> <chr> <int> <dbl>
## 1 1880 F Mary 7065 0.0724
## 2 1880 F Anna 2604 0.0267
## 3 1880 F Emma 2003 0.0205
## 4 1880 F Elizabeth 1939 0.0199
## 5 1880 F Minnie 1746 0.0179
## 6 1880 F Margaret 1578 0.0162
## 7 1880 F Ida 1472 0.0151
## 8 1880 F Alice 1414 0.0145
## 9 1880 F Bertha 1320 0.0135
## 10 1880 F Sarah 1288 0.0132
## # ℹ 1,924,655 more rows4.4.2 Exploring data
Filtering and arranging for one year
babynames %>%
# Filter for the year 1990
filter(year == 1990) %>%
# Sort the number column in descending order
arrange(desc(n))## # A tibble: 24,719 × 5
## year sex name n prop
## <dbl> <chr> <chr> <int> <dbl>
## 1 1990 M Michael 65282 0.0303
## 2 1990 M Christopher 52332 0.0243
## 3 1990 F Jessica 46475 0.0226
## 4 1990 F Ashley 45558 0.0222
## 5 1990 M Matthew 44800 0.0208
## 6 1990 M Joshua 43216 0.0201
## 7 1990 F Brittany 36538 0.0178
## 8 1990 F Amanda 34408 0.0168
## 9 1990 M Daniel 33815 0.0157
## 10 1990 M David 33742 0.0157
## # ℹ 24,709 more rowsFinding the most popular names each year
babynames %>%
# Find the most common name in each year
group_by(year) %>%
slice_max(n, n = 1)## # A tibble: 138 × 5
## # Groups: year [138]
## year sex name n prop
## <dbl> <chr> <chr> <int> <dbl>
## 1 1880 M John 9655 0.0815
## 2 1881 M John 8769 0.0810
## 3 1882 M John 9557 0.0783
## 4 1883 M John 8894 0.0791
## 5 1884 M John 9388 0.0765
## 6 1885 F Mary 9128 0.0643
## 7 1886 F Mary 9889 0.0643
## 8 1887 F Mary 9888 0.0636
## 9 1888 F Mary 11754 0.0620
## 10 1889 F Mary 11648 0.0616
## # ℹ 128 more rowsVisualizing names with ggplot2
selected_names <- babynames %>%
# Filter for the names Steven, Thomas, and Matthew. And female only
filter(name %in% c("Steven", "Thomas", "Matthew") & sex == "F")
# Plot the names using a different color for each name
ggplot(selected_names, aes(x = year, y = n, color = name)) +
geom_line()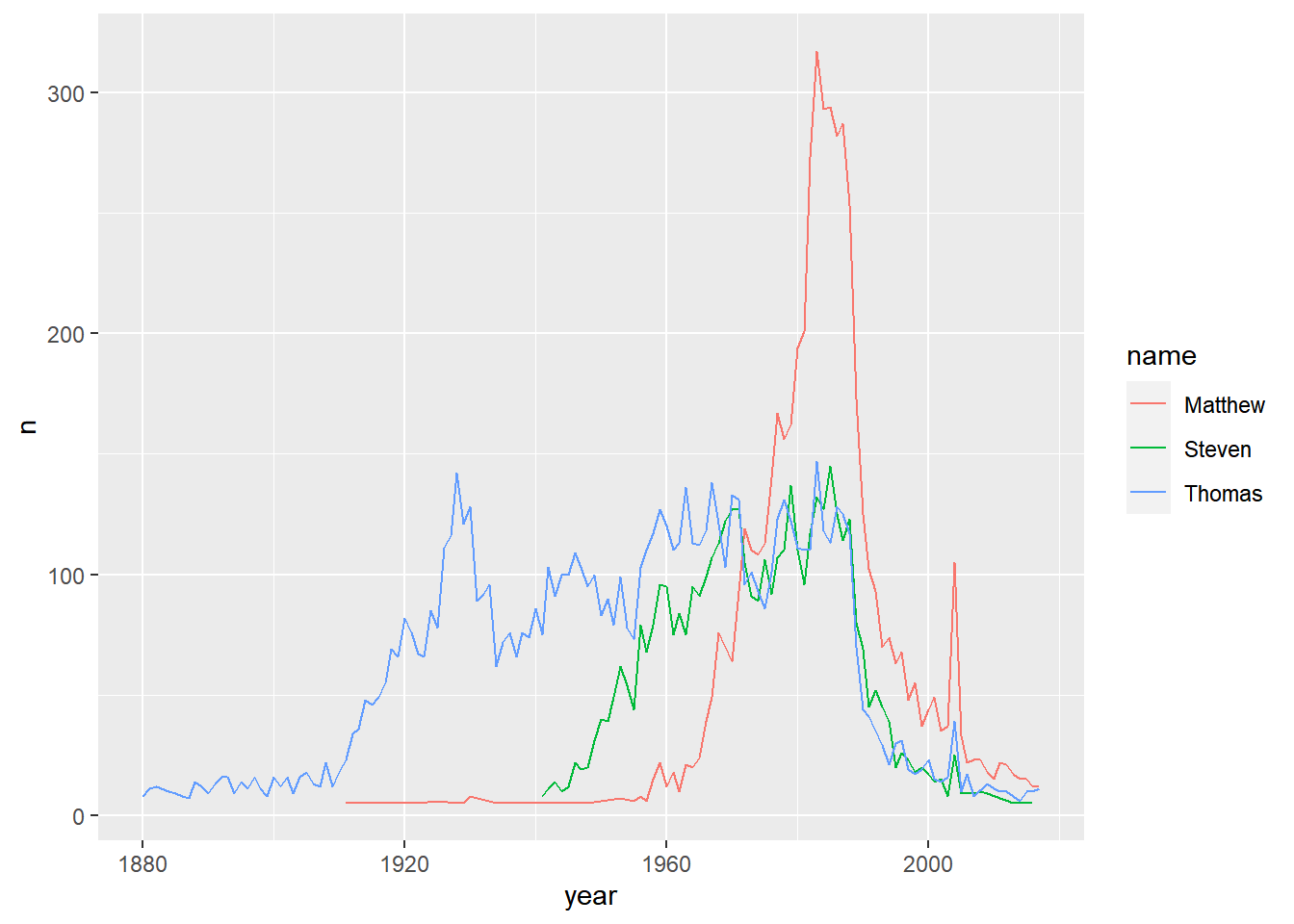
Finding the year each name is most common
# Calculate the fraction of people born each year with the same name
babynames %>%
group_by(year) %>%
mutate(year_total = sum(n)) %>%
ungroup() %>%
mutate(fraction = n / year_total) %>%
# Find the year each name is most common
group_by(name) %>%
slice_max(fraction, n = 1)## # A tibble: 97,351 × 7
## # Groups: name [97,310]
## year sex name n prop year_total fraction
## <dbl> <chr> <chr> <int> <dbl> <int> <dbl>
## 1 2014 M Aaban 16 0.00000783 3696311 0.00000433
## 2 2014 F Aabha 9 0.00000461 3696311 0.00000243
## 3 2016 M Aabid 5 0.00000248 3652968 0.00000137
## 4 2016 M Aabir 5 0.00000248 3652968 0.00000137
## 5 2016 F Aabriella 11 0.0000057 3652968 0.00000301
## 6 2015 F Aada 5 0.00000257 3688687 0.00000136
## 7 2015 M Aadam 22 0.0000108 3688687 0.00000596
## 8 2009 M Aadan 23 0.0000108 3815638 0.00000603
## 9 2014 M Aadarsh 18 0.0000088 3696311 0.00000487
## 10 2009 M Aaden 1267 0.000598 3815638 0.000332
## # ℹ 97,341 more rowsAdding the total and maximum for each name
You’ll divide each name by the maximum for that name. This means that every name will peak at 1.
babynames %>%
# Add columns name_total and name_max for each name
group_by(name) %>%
mutate(name_total = sum(n),
name_max = max(n)) %>%
# Ungroup the table
ungroup() %>%
# Add the fraction_max column containing the number by the name maximum
mutate(fraction_max = n / name_max)## # A tibble: 1,924,665 × 8
## year sex name n prop name_total name_max fraction_max
## <dbl> <chr> <chr> <int> <dbl> <int> <int> <dbl>
## 1 1880 F Mary 7065 0.0724 4138360 73982 0.0955
## 2 1880 F Anna 2604 0.0267 891245 15666 0.166
## 3 1880 F Emma 2003 0.0205 655629 22704 0.0882
## 4 1880 F Elizabeth 1939 0.0199 1634860 20744 0.0935
## 5 1880 F Minnie 1746 0.0179 159562 3274 0.533
## 6 1880 F Margaret 1578 0.0162 1250392 28466 0.0554
## 7 1880 F Ida 1472 0.0151 186955 4450 0.331
## 8 1880 F Alice 1414 0.0145 562006 11956 0.118
## 9 1880 F Bertha 1320 0.0135 208550 5051 0.261
## 10 1880 F Sarah 1288 0.0132 1077215 28484 0.0452
## # ℹ 1,924,655 more rowsVisualizing the normalized change in popularity
You picked a few names and calculated each of them as a fraction of their peak. This is a type of “normalizing” a name, where you’re focused on the relative change within each name rather than the overall popularity of the name.
names_normalized <- babynames %>%
group_by(name) %>%
mutate(name_total = sum(n),
name_max = max(n)) %>%
ungroup() %>%
mutate(fraction_max = n / name_max)
names_filtered <- names_normalized %>%
# Filter for the names Steven, Thomas, and Matthew. And male only
filter(name %in% c("Steven", "Thomas", "Matthew") & sex == "M")
# Visualize these names over time
ggplot(names_filtered, aes(x = year, y = fraction_max, color = name)) +
geom_line()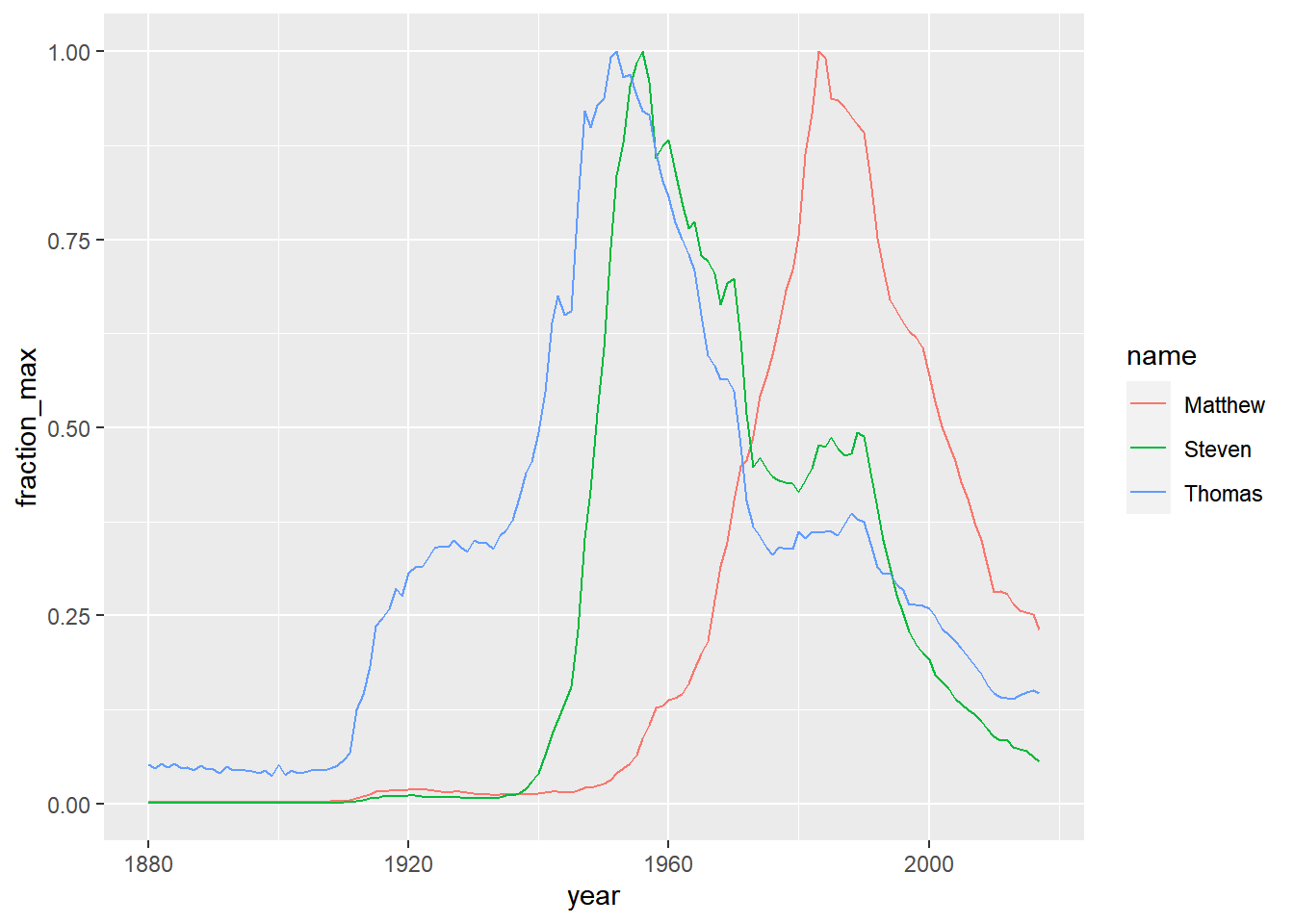
As you can see, the line for each name hits a peak at 1, although the peak year differs for each name.
Using ratios to describe the frequency of a name
Window function: lag()
v <- c(1, 3, 6, 14); v
## 1 3 6 14
lag(v)
## NA 1 3 6
v - lag(v)
## NA 2 3 8Notice that the first observation for each name is missing a ratio, since there is no previous year.
babynames_fraction <- babynames %>%
group_by(year) %>%
mutate(year_total = sum(n)) %>%
ungroup() %>%
mutate(fraction = n / year_total)
babynames_fraction %>%
# Arrange the data in order of name, then year
arrange(name, year) %>%
# Group the data by name
group_by(name) %>%
# Add a ratio column that contains the ratio of fraction between each year
mutate(ratio = fraction / lag(fraction))## # A tibble: 1,924,665 × 8
## # Groups: name [97,310]
## year sex name n prop year_total fraction ratio
## <dbl> <chr> <chr> <int> <dbl> <int> <dbl> <dbl>
## 1 2007 M Aaban 5 0.00000226 3994007 0.00000125 NA
## 2 2009 M Aaban 6 0.00000283 3815638 0.00000157 1.26
## 3 2010 M Aaban 9 0.00000439 3690700 0.00000244 1.55
## 4 2011 M Aaban 11 0.00000542 3651914 0.00000301 1.24
## 5 2012 M Aaban 11 0.00000543 3650462 0.00000301 1.00
## 6 2013 M Aaban 14 0.00000694 3637310 0.00000385 1.28
## 7 2014 M Aaban 16 0.00000783 3696311 0.00000433 1.12
## 8 2015 M Aaban 15 0.00000736 3688687 0.00000407 0.939
## 9 2016 M Aaban 9 0.00000446 3652968 0.00000246 0.606
## 10 2017 M Aaban 11 0.0000056 3546301 0.00000310 1.26
## # ℹ 1,924,655 more rowsBiggest jumps in a name
To look further into the names that experienced the biggest jumps in popularity in consecutive years.
babynames_ratios_filtered <- babynames_fraction %>%
arrange(name, year) %>%
group_by(name) %>%
mutate(ratio = fraction / lag(fraction)) %>%
filter(fraction >= 0.00001)
babynames_ratios_filtered %>%
# Extract the largest ratio from each name
slice_max(ratio, n = 1) %>%
# Sort the ratio column in descending order
arrange(desc(ratio)) %>%
# Filter for fractions greater than or equal to 0.001
filter(fraction >= 0.001)## # A tibble: 529 × 8
## # Groups: name [529]
## year sex name n prop year_total fraction ratio
## <dbl> <chr> <chr> <int> <dbl> <int> <dbl> <dbl>
## 1 2010 F Sophia 20639 0.0105 3690700 0.00559 2371.
## 2 2001 F Olivia 13978 0.00706 3741451 0.00374 2352.
## 3 2002 F Isabella 12166 0.00616 3736042 0.00326 2031.
## 4 2014 M Joseph 12106 0.00592 3696311 0.00328 2018.
## 5 2016 F Emma 19471 0.0101 3652968 0.00533 1966.
## 6 2016 F Elizabeth 9542 0.00495 3652968 0.00261 1927.
## 7 2013 M Luke 9568 0.00474 3637310 0.00263 1914.
## 8 2011 M Samuel 11340 0.00559 3651914 0.00311 1890
## 9 2015 F Evelyn 9358 0.00481 3688687 0.00254 1875.
## 10 2010 M Isaac 9354 0.00456 3690700 0.00253 1871.
## # ℹ 519 more rows